Review Article
Dysregulation of Protein Synthesis and its Implications for CNS Disorders: Detection Methods, Diseases and Therapy an Update
Kiran Kumar H.B, K. Ramachandra Kini, Kamatchi. K, Ranjini P.
Licence:
Attribution-Non-commercial 4.0 International (CC BY-NC 4.0)This license enables reusers to distribute, remix, adapt, and build upon the material in any medium or format for noncommercial purposes only, and only so long as attribution is given to the creator.
Indian Journal of Genetics and Molecular Research 14(2):p 53-65, July-Dec 2025. | DOI: 10.21088/ijgmr.2319.4782.14225.2
How Cite This Article:
Kiran Kumar HB, Kini KR, Kamatachi C, et al. Dysregulation of protein synthesis and its implications for CNS disorders: detection methods, diseases and therapy an update. Ind J Genet Mol Res. 2025;14(2):53-65.Timeline
Abstract
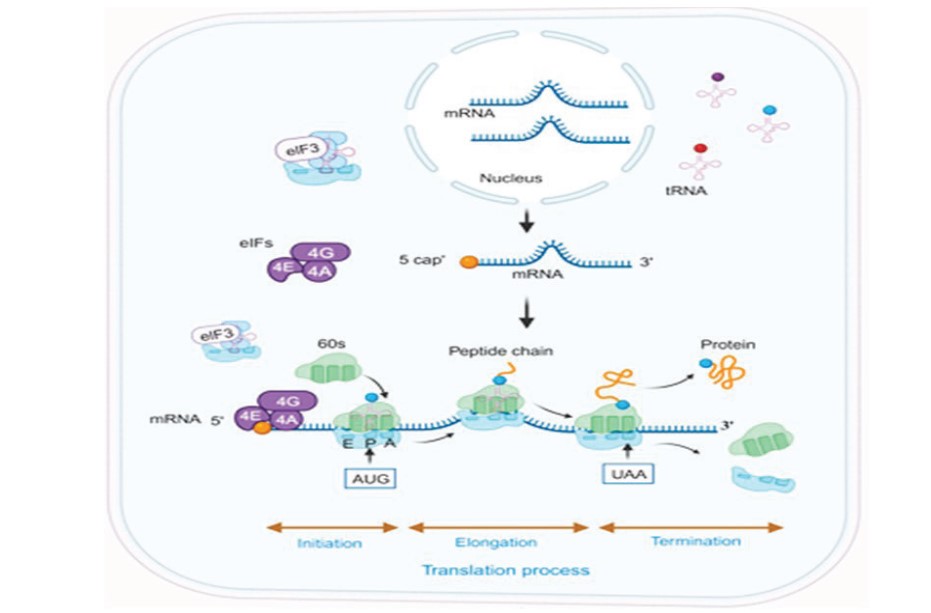
Protein translation is central to gene expression in the central nervous system (CNS) because it is the crucial process of converting messenger RNA (mRNA) into functional proteins, which are vital for the structure, function, and regulation of neurons and their circuits. Regulation of protein synthesis involves posttranscriptional modifications and coordination between transcription and mRNA turnover to enable rapid signaling and gene expression changes in cells of CNS. This process is highly regulated and essential for key CNS functions such as synaptic plasticity, learning, and memory. Impaired translation is linked to several CNS disorders. The present review covers basic areas of gene-regulation and deregulation to highlight the unique features of the mechanism and their relevance to CNS. Further, it updates on the current methods of detection, and CNS disorders with details of the mechanisms and proteins, pathways involved in brief. Finally, the potential of targeting the proteins and intermediates of the mechanism as therapeutic targets is discussed. In the post-Human genome sequencing era CNS disorders are implicated as major source of Human disorders. Novel methods to treat these disorders are need of the hour, thus the detail study of protein synthesis at all levels would enable positive leads towards this direction. The present review is a brief update of the current literature in this important area of biomedical research.
References
- 1. Cell and Molecular Biology.De Robertis, 8th Edition 2020.Lww Rs Pharmacy Exclusive (Cbs).
- 2. Molecular Biology of the Cell, Bruce Alberts, Rebecca Heald, et al.7th Edition. 2022 .Norton & Co.
- 3. Olga M. Alekhina,Ilya M. Terenin, ,Sergey E. Dmitriev and Konstantin S.Vassilenko. Functional Cyclization of Eukaryotic mRNAs. Int. J. Mol. Sci. 2020, 21(5), 1677; https://doi. org/10.3390/ijms21051677.
- 4. Raphael Böhm,Stefan Imseng, Roman P. JakobMichael N. Hall, Timm Maier, Sebastian Hiller. The dynamic mechanism of 4E-BP1 recognition and phosphorylation by mTORC1. Molecular Cell.Volume 81, Issue 11, 3 June 2021, Pages 2403-2416.e5.
- 5. Hyung Don Ryoo.The integrated stress response in metabolic adaptation Journal of Biological Chemistry.Volume 300, Issue 4, April 2024, 107151.
- 6. Paula Llabata, Julia Richte, Isabel Faus, Karolina Sska-Durdasiak, Lukas Hubert Zeh, Jose Gadea, Marie-Theres Hauser. Involvement of the eIF2α Kinase GCN2 in UV-B Responses. Front. Plant Sci., 28 November 2019.Sec. Plant Abiotic Stress.Volume 10 - 2019 | https://doi. org/10.3389/fpls.2019.01492.
- 7. Neethi Nandagopal, Philippe P Roux. Regulation of global and specific mRNA translation by the mTOR signaling pathway Translation (Austin). 2015 Feb 2;3(1):e983402. doi: 10.4161/21690731.2014.983402.
- 8. Kathrin Leppek, Rhiju Da, Maria Barna. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them Nat Rev Mol Cell Biol. 2017 Nov 22;19(3):158–174. doi: 10.1038/nrm.2017.103
- 9. Joanna Somers,Tuija Pöyry, Anne E. Willis.A perspective on mammalian upstream open reading frame function International Journal of Biochemistry & Cell Biology.Volume 45, Issue 8, August 2013, Pages 1690-1700.
- 10. Lauren Endres, Peter C Dedon, Thomas J BegleyCodon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. Review RNA Biol. 2015;12(6):603-14. doi: 10.1080/15476286.2015.1031947.
- 11. Hui-Jun Dong, Rui Zhang, Yu Kuang, XiaoJia Wang. Selective regulation in ribosome biogenesis and protein production for efficient viral translation.Arch Microbiol. 2020 Oct 29;203(3):1021–1032. doi: 10.1007/s00203-020- 02094-5.
- 12. Yanyan Gao, Linlin Guo, Gaoxiang Shi, Ruifang Wang, Xu’an Wang, Jizhong Lou. Emerging roles of ribosome translation in stem cells and stem cell therapy - a review Cell Biosci. 2025 May 28;15:71. doi: 10.1186/s13578- 025-01412-y.
- 13. Kate D Meyer, Deepak P Patil, Jun Zhou, Alexandra Zinoviev, Maxim A Skabkin, Olivier Elemento, Tatyana V Pestova, ShuBing Qian, Samie R Jaffrey.5′ UTR m6A Promotes Cap-Independent Translation Cell. 2015 Oct 22;163(4):999–1010. doi: 10.1016/j. cell.2015.10.012.
- 14. Amy SY Lee, Philip J Kranzusch, Jennifer A Doudna, Jamie HD Cate.eIF3d is an mRNA cap-binding protein required for specialized translation initiation Nature. 2016 Aug 4;536(7614):96–99. doi: 10.1038/nature18954.
- 15. Yun Yang, Zefeng Wang, IRES-mediated cap-independent translation, a path leading to hidden proteome, Journal of Molecular Cell Biology, Volume 11, Issue 10, October 2019, Pages 911–919, https://doi.org/10.1093/ jmcb/mjz091.
- 16. Dennis H. Bamford and Mark Zuckerman. Encyclopedia of Virology. Fourth Edition • Academic Press.2021.
- 17. Song, P., Yang, F., Jin, H. et al. The regulation of protein translation and its implications for cancer. Sig Transduct Target Ther 6, 68 (2021). https://doi.org/10.1038/s41392-020-00444-9.
- 18. Esteban A Orellana, Elisabeth Siegal, Richard I Gregory.tRNA dysregulation and disease Nat Rev Genet. 2022 Jun 9;23(11):651–664. doi: 10.1038/s41576-022-00501-9.
- 19. Zsofia Turi, Matthew Lacey, Martin Mistrik, Pavel Moudry.Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging Aging (Albany NY). 2019 Apr 26;11(8):2512– 2540. doi: 10.18632/aging.101922.
- 20. PanelIlias Georgakopoulos-Soares, Guillermo E. Parada, Martin Hemberg.Secondary structures in RNA synthesis, splicing and translation Computational and Structural Biotechnology Journal Volume 20, 2022, Pages 2871-2884.
- 21. Jacob O’Brien, Heyam Hayder, Yara Zayed, Chun Peng. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation Front Endocrinol (Lausanne). 2018 Aug 3;9:402. doi: 10.3389/fendo.2018.00402.
- 22. Géssica C Barros, Sofia Guerrero, Gustavo M Silva. The Central Role of Translation Elongation in Response to Stress Biochem Soc Trans. 2023 Jun 28;51(3):959–969. doi: 10.1042/ BST20220584.
- 23. Anna R Guzikowski, Alex T Harvey, Jingxiao Zhang, Shihui Zhu, Kyle Begovich, Molly H Cohn, James E Wilhelm, Brian M Zid.Differential translation elongation directs protein synthesis in response to acute glucose deprivation in yeast. RNA Biol. 2022 May 1;19(1):636–649. doi: 10.1080/15476286.2022.2065784.
- 24. Natalia Shcherbik, Dimitri G Pestov. The Impact of Oxidative Stress on Ribosomes: From Injury to Regulation.Cells. 2019 Nov 2;8(11):1379. doi: 10.3390/cells8111379.
- 25. Marina V Rodnina. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci. 2016 Jun 8;25(8):1390–1406. doi: 10.1002/pro.2950.
- 26. Qiu Peng, Yujuan Zhou, Linda Oyang, Nayiyuan Wu, Yanyan Tang, Min Su, Xia Luo, Ying Wang, Xiaowu Sheng, Jian Ma, Qianjin Liao. Impacts and mechanisms of alternative mRNA splicing in cancer metabolism, immune response, and therapeutics. Mol Ther. 2021 Nov 15;30(3):1018–1035. doi: 10.1016/j. ymthe.2021.11.010.
- 27. Xumin Ou, Jingyu Cao, Anchun Cheng, Maikel P Peppelenbosch, Qiuwei Pan .Errors in translational decoding: tRNA wobbling or misincorporation? PLoS Genet. 2019 Mar 28;15(3):e1008017. doi: 10.1371/journal. pgen.1008017.
- 28. Madhuparna Pandit,Md Noor Akhtar, Susinder Sundaram,Sarthak Sahoo,Lekha E. Manjunath, Sandeep M. Eswarappa.Termination codon readthrough of NNAT mRNA regulates calcium-mediated neuronal differentiation Journal home page for Journal of Biological Chemistry.Volume 299, Issue 9, September 2023, 105184.
- 29. Caleb M Embree, Rabab Abu-Alhasan, Guramrit Singh. Features and factors that dictate if terminating ribosomes cause or counteract nonsense-mediated mRNA decay.J Biol Chem. 2022 Oct 13;298(11):102592. doi: 10.1016/j.jbc.2022.102592.
- 30. F. Lejeune.Nonsense-mediated mRNA decay at the crossroads of many cellular pathways. BMB Reports, 50 (4) (2017), pp. 175-185, 10.5483/BMBRep.2017.50.4.015.
- 31. Dabrowski, M., Bukowy-Bieryllo, Z., & Zietkiewicz, E. (2015). Translational readthrough potential of natural termination codons in eucaryotes – The impact of RNA sequence. RNA Biology, 12(9), 950–958. https://doi.org/10.1080/15476286.2015. 1068497.
- 32. Andrew G Cridge , Caillan Crowe-McAuliffe , Suneeth F Mathew , Warren P TateEukaryotic translational termination efficiency is influenced by the 3′ nucleotides within the ribosomal mRNA channel. Nucleic Acids Research, Volume 46, Issue 4, 28 February 2018, Pages 1927–1944, https://doi.org/10.1093/ nar/gkx1315.
- 33. Paul F Agris, Amithi Narendran, Kathryn Sarachan, Ville YP Väre, Emily Eruysal. The Role of RNA Modifications in Translational Fidelity.Enzymes. 2017 Apr 22;41:1–50. doi: 10.1016/bs.enz.2017.03.005.
- 34. J.W. Drysdale, H.N. Munro.Polysome profiles obtained from mammalian tissues by an improved procedure.Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis Volume 138, Issue 3, 30 May 1967, Pages 616-618.
- 35. Helen A. King, André P. Gerber.Translatome profiling: methods for genome-scale analysis of mRNA translation. Briefings in Functional Genomics, Volume 15, Issue 1, January 2016, Pages 22–31, https://doi.org/10.1093/bfgp/ elu045.
- 36. Shuo Liang, Hermano Martins Bellato, Julie Lorent, Fernanda C S Lupinacci, Christian Oertlin, Vincent van Hoef, Victor P Andrade, Martín Roffé, Laia Masvidal, Glaucia N M Hajj, Ola Larsson, Polysome-profiling in small tissue samples, Nucleic Acids Research, Volume 46, Issue 1, 9 January 2018, Page e3, https://doi. org/10.1093/nar/gkx940.
- 37. Nicholas T Ingolia, Gloria A Brar, Noam Stern-Ginossar, Michael S Harris, Gaëlle J S Talhouarne, Sarah E Jackson, Mark R Wills, Jonathan S Weissman. Ribosome Profiling Reveals Pervasive Translation Outside of Annotated Protein-Coding Genes.Cell Rep. 2014 Aug 21;8(5):1365–1379. doi: 10.1016/j. celrep.2014.07.045.
- 38. Nicholas T Ingolia.Ribosome Footprint Profiling of Translation throughout the Genome Cell. 2016 Mar 24;165(1):22–33. doi: 10.1016/j.cell.2016.02.066.
- 39. Piyada Juntawong,Thomas Girke, Jérémie Bazin, Julia Bailey-Serres. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci U S A. 2013 Dec 23;111(1):E203– E212. doi: 10.1073/pnas.1317811111.
- 40. Toshifumi Inada, Eric Winstall, Salvador Z Tarun Jr, John R Yates 3rd, Dave Schieltz, Alan B Sachs. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs RNA. 2002 Jul;8(7):948-58. doi: 10.1017/s1355838202026018.
- 41. Shifeng Xue, Maria Barna. Specialized ribosomes: a new frontier in gene regulation and organismal biology Nat Rev Mol Cell Biol. 2012 May 23;13(6):355–369. doi: 10.1038/ nrm3359.
- 42. Alison Barbara Ross, Julian David Langer, Marko Jovanovic. Proteome Turnover in the Spotlight: Approaches, Applications, and Perspectives. Molecular & Cellular Proteomics. Volume 20, 2021, 100016.
- 43. Yuming Jiang, Devasahayam Arokia,Balaya Rex, Dina Schuster,Benjamin A. Neely, Germán L. Rosano,Norbert Volkmar, Amanda Momenzadeh, Trenton M. Peters-Clarke,Susan B. Egbert, Simion Kreimer, Emma H. Doud, Oliver M. Crook, Amit Kumar Yadav, Muralidharan Vanuopadath, Adrian D. Hegeman,Martín L. Mayta, Anna G. Duboff, Nicholas M. Riley, Robert L. Moritz,Jesse G. Meyer. Comprehensive Overview of Bottom Up Proteomics Using Mass Spectrometry. ACS Measurement Science AuVol 4/Issue 4 Article ReviewJune 4, 2024.
- 44. Apostolia M. Tsimberidou, Elena Fountzilas, Leonidas Bleris, Razelle Kurzrock,Transcriptomics and solid tumors: The next frontier in precision cancer medicine. Seminars in Cancer Biology, Volume 84,2022. Review.
- 45. Michael VanInsberghe, Jeroen van den Berg, Amanda Andersson-Rolf, Hans Clevers, Alexander van Oudenaarden. Single-cell Riboseq reveals cell cycle-dependent translational pausing. Nature. 2021 Sep;597(7877):561-565. doi: 10.1038/s41586-021-03887-4. Epub 2021 Sep 8.
- 46. An Zhou, Fang Bian.Editorial: Proteostasis in central nervous system disorders.Front Mol Neurosci. 2024 Mar 18;17:1394171. doi: 10.3389/fnmol.2024.1394171.
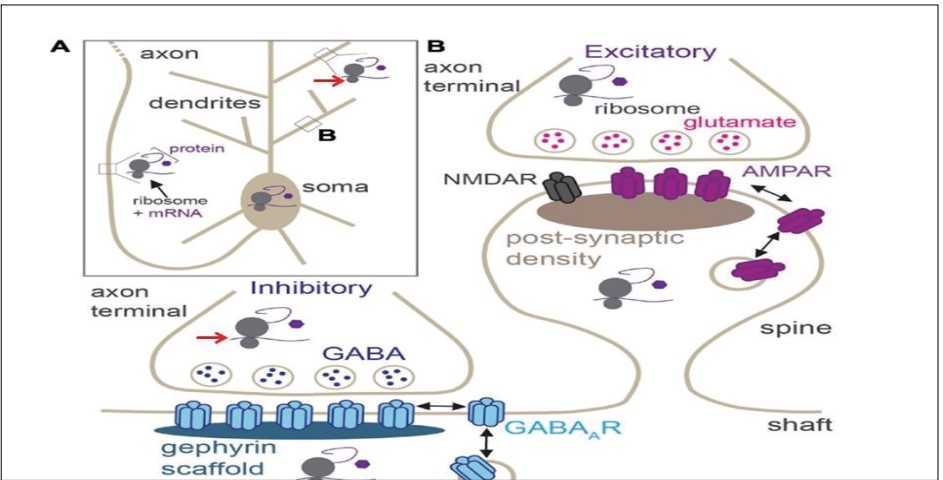
- 47. Sulagna Das, Maria Vera, Valentina Gandin, Robert H Singer, Evelina Tutucci. Intracellular mRNA transport and localized translation.Nat Rev Mol Cell Biol. 2021 Apr 9;22(7):483–504. doi: 10.1038/s41580-021-00356-8.
- 48. Dipen Rajgor,Theresa M. Welle,Katharine R. Smith,Katharine R. Smith.The Coordination of Local Translation, Membranous Organelle Trafficking, and Synaptic Plasticity in Neurons. Front. Cell Dev. Biol., 14 July 2021.Sec. Membrane Traffic and Organelle Dynamics. Volume 9 - 2021 | https://doi.org/10.3389/ fcell.2021.711446.
- 49. Stephanie L Moon, Roy Parker.EIF2B2 mutations in vanishing white matter disease hypersuppress translation and delay recovery during the integrated stress response RNA. 2018 Jun;24(6):841–852. doi: 10.1261/ rna.066563.118.
- 50. Marjo S van der Knaap, Anne Fogli, Odile Boespflug-Tanguy, Truus EM Abbink, and Raphael Schiffmann. Childhood Ataxia with Central Nervous System Hypomyelination / Vanishing White Matter. GeneReviews® [Internet].Seattle (WA): University of Washington, Seattle; 1993-2025. Synonyms: CACH/VWM, Leukoencephalopathy with Vanishing White Matter.
- 51. Rebecca Meyer-Schuman, Anthony Antonellis. Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease Hum Mol Genet. 2017 Jun 15;26(R2):R114–R127. doi: 10.1093/ hmg/ddx231.
- 52. Faith C J Davies, Jilly E Hope, Fiona McLachlan, Francis Nunez, Jennifer Doig , Hemant Bengani, Colin Smith, Catherine M Abbott. Biallelic mutations in the gene encoding eEF1A2 cause seizures and sudden death in F0 mice Sci Rep. 2017 Apr 5;7:46019. doi: 10.1038/ srep46019.
- 53. Eileen Chen, Simpson Joseph.Fragile X Mental Retardation Protein: A Paradigm for Translational Control by RNA-Binding Proteins.Biochimie. Author manuscript; available in PMC: 2016 Jul 1.
- 54. Rita Marques, Rafaela Lacerda, Luísa Romão. Internal Ribosome Entry Site (IRES)-Mediated Translation and Its Potential for Novel mRNABased Therapy Development Biomedicines. 2022 Aug 2;10(8):1865. doi: 10.3390/ biomedicines10081865.
- 55. Seyed Khalil Rashidi, Ata Kalirad, Shahram Rafie, Ebrahim Behzad , Mitra Ansari Dezfouli. The role of microRNAs in neurobiology and pathophysiology of the hippocampus Front Mol Neurosci. 2023 Sep 4;16:1226413. doi: 10.3389/fnmol.2023.1226413.
- 56. Xiaoyu Dong, Shuyan Cong.MicroRNAs in Huntington’s Disease: Diagnostic Biomarkers or Therapeutic Agents? Front Cell Neurosci. 2021 Aug 6;15:705348. doi: 10.3389/ fncel.2021.705348.
- 57. Long Wang, Xindong Shui, Yuelin Diao, Duoting Chen, Ying Zhou, Tae Ho Lee. Potential Implications of miRNAs in the Pathogenesis, Diagnosis, and Therapeutics of Alzheimer’s Disease. Int J Mol Sci. 2023 Nov 13;24(22):16259. doi: 10.3390/ijms242216259.
- 58. Nicola Lopizzo, Valentina Zonca, Nadia Cattane, Carmine Maria Pariante , Annamaria Cattaneo.miRNAs in depression vulnerability and resilience: novel targets for preventive strategies J Neural Transm (Vienna). 2019 Jul 26;126(9):1241–1258. doi: 10.1007/s00702-019- 02048-2.
- 59. Julie Meffre, Séverine Chaumont-Dubel, Clotilde Mannoury la Cour, Florence Loiseau, David J G Watson, Anne Dekeyne, Martial Séveno, Jean-Michel Rivet, Florence Gaven, Paul Déléris, Denis Hervé, Kevin C F Fone, Joël Bockaert, Mark J Millan, Philippe Marin.5-HT6 receptor recruitment of mTOR as a mechanism for perturbed cognition in schizophrenia EMBO Mol Med. 2012 Oct 2;4(10):1043–1056. doi: 10.1002/emmm.201201410.
- 60. Anand Gururajan, Maarten van den Buuse. Is the mTOR-signalling cascade disrupted in Schizophrenia?.J. Neurochem. (2014) 129, 377– 387.
- 61. Henry Paulson. Repeat expansion diseases. Handb Clin Neurol. 2018;147:105–123. doi: 10.1016/B978-0-444-63233-3.00009-9.
- 62. Lindsey D Goodman, Nancy M Bonini.Repeatassociated non-AUG (RAN) translation mechanisms running into focus for GGGGCCrepeat associated ALS/FTD. Prog Neurobiol. 2019 Sep 21;183:101697. doi: 10.1016/j. pneurobio.2019.101697.
- 63. Michael G Kearse, Jeremy E Wilusz. Non-AUG translation: a new start for protein synthesis in eukaryotes.Genes Dev. 2017 Sep 1;31(17):1717– 1731. doi: 10.1101/gad.305250.117.
- 64. Monica Banez-Coronel, Laura P.W. Ranum. Repeat-associated non-AUG (RAN) translation: insights from pathology. Laboratory Investigation. Volume 99, Issue 7, July 2019, Pages 929-942. Nat Commun. 2018 Jan 4;9:51. doi: 10.1038/s41467-017-02495-z.
- 65. Weiwei Cheng, Shaopeng Wang, Alexander A Mestre, Chenglai Fu, Andres Makarem, Fengfan Xian, Lindsey R Hayes, Rodrigo Lopez-Gonzalez, Kevin Drenner, Jie Jiang, Don W Cleveland, Shuying Sun. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation Nat Commun. 2018 Jan 4;9:51. doi: 10.1038/s41467-017-02495-z.
- 66. Anne Cammas, Stefania Millevoi, Weiwei Cheng, Shaopeng Wang, Alexander A Mestre, Chenglai Fu, Andres Makarem, Fengfan Xian, Lindsey R Hayes, Rodrigo Lopez-Gonzalez, Kevin Drenner, Jie Jiang , Don W Cleveland , Shuying Sun.RNA G-quadruplexes: emerging mechanisms in disease.Nucleic Acids Res. 2016 Dec 24;45(4):1584–1595. doi: 10.1093/nar/ gkw1280.
- 67. Marzena Wojciechowska, Marta Olejniczak, Paulina Galka-Marciniak,Magdalena Jazurek and Wlodzimierz J. Krzyzosiak. RAN translation and frameshifting as translational challenges at simple repeats of human neurodegenerative disorders. Nucleic Acids Research, 2014, Vol. 42, No. 19 11849–11864 doi: 10.1093/nar/gku794.
- 68. Heleen M van ‘t Spijker, Sandra Almeida. How Villains are Made: The Translation of Dipeptide Repeat Proteins in C9ORF72- ALS/FTD.Gene. 2023 Jan 6;858:147167. doi: 10.1016/j.gene.2023.147167.
- 69. Ilias Georgakopoulos-Soares, Guillermo E Parada, Martin Hemberg.structures in RNA synthesis, splicing and translation. Comput Struct Biotechnol J. 2022 May 27;20:2871–2884. doi: 10.1016/j.csbj.2022.05.041.
- 70. John R P Knight, Gavin Garland, Tuija Pöyry, Emma Mead, Nikola Vlahov, Aristeidis Sfakianos, Stefano Grosso, Fabio De-LimaHedayioglu, Giovanna R Mallucci, Tobias von der Haar, C Mark Smales, Owen J Sansom, Anne E Willis. Control of translation elongation in health and disease. Dis Model Mech. 2020 Mar 26;13(3):dmm043208. doi: 10.1242/dmm.043208.
- 71. Xin Wang, Qian Yang, Xueyan Zhou, C Dirk Keene, Alexey G Ryazanov, Tao Ma .Suppression of eEF2 phosphorylation alleviates synaptic failure and cognitive deficits in mouse models of Down syndrome Alzheimers Dement. 2024 Jun 27;20(8):5357– 5374. doi: 10.1002/alz.13916.
- 72. Zizheng Dong, Jian-Ting Zhang.EIF3 p170, a Mediator of Mimosine Effect on Protein Synthesis and Cell Cycle Progression This is the final version - click for previous version Molecular Biology of the Cell Vol. 14, No. 9.
- 73. Heinz-Josef Klümpen, Jos H Beijnen, Howard Gurney, Jan HM. Schellens. Inhibitors of mTOR Oncologist. 2010 Dec 8;15(12):1262–1269. doi: 10.1634/theoncologist.2010-0196.
- 74. Weiwei Cheng, Shaopeng Wang, Alexander A Mestre, Chenglai Fu, Andres Makarem, Fengfan Xian, Lindsey R Hayes, Rodrigo Lopez-Gonzalez, Kevin Drenner, Jie Jiang, Don W Cleveland, Shuying Sun. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat Commun. 2018 Jan 4;9:51. doi: 10.1038/s41467-017-02495-z.
- 75. Jeffrey P MacKeigan, Darcy A Krueger. Differentiating the mTOR inhibitors everolimus and sirolimus in the treatment of tuberous sclerosis complex. Neuro Oncol. 2015 Aug 19;17(12):1550–1559. doi: 10.1093/ neuonc/nov152.
- 76. Tramutola, A., Lanzillotta, C., and Di Domenico, F. (2017). Targeting mTOR to reduce Alzheimer-related cognitive decline: from current hits to future therapies. Expert Rev. Neurother. 17 (1), 33–45. doi:10.1080/147 37175.2017.1244482.
- 77. Ricardo Moreno, Javier Recio, Santiago Barber, Carmen Gil, Ana Martinez. The emerging role of mixed lineage kinase 3 (MLK3) and its potential as a target for neurodegenerative diseases therapies. Review Eur J Med Chem. 2023 Sep 5:257:115511. doi: 10.1016/j. ejmech.2023.115511. Epub 2023 May 24.
- 78. Ashwini Saxena, Giselli Scaini, Daniela V Bavaresco, Camila Leite, Samira S Valvassoria, André F Carvalho, João Quevedo. Role of Protein Kinase C in Bipolar Disorder: A Review of the Current Literature. Mol Neuropsychiatry. 2017 Oct 7;3(2):108–124. doi: 10.1159/000480349.
- 79. Ling-Zhi Xu, Bing-Qiu Li, Jian-Ping Jia. DAPK1: a Novel Pathology and Treatment Target for Alzheimer’s Disease. Mol Neurobiol. 2019 Apr;56(4):2838-2844. doi: 10.1007/s12035- 018-1242-2. Epub 2018 Jul 31.
Data Sharing Statement
Funding
Author Contributions
Ethics Declaration
Acknowledgements
Conflicts of Interest
About this article
Cite this article
Kiran Kumar HB, Kini KR, Kamatachi C, et al. Dysregulation of protein synthesis and its implications for CNS disorders: detection methods, diseases and therapy an update. Ind J Genet Mol Res. 2025;14(2):53-65.
Licence:
Attribution-Non-commercial 4.0 International (CC BY-NC 4.0)This license enables reusers to distribute, remix, adapt, and build upon the material in any medium or format for noncommercial purposes only, and only so long as attribution is given to the creator.
| Received | Accepted | Published |
|---|---|---|
| August 25, 2025 | September 24, 2025 | December 28, 2025 |
DOI: 10.21088/ijgmr.2319.4782.14225.2
Keywords
Ribosome ProfilingAminoacyl-tRNA Synthetases (ARSs)Repeat-Associated Non-Aug (Ran) TranslationSearch for Similar Articles
Similar Articles
Article Level Metrics
Last UpdatedSaturday 07 February 2026, 08:54:31 (IST)
Accesses
Citations
Download citation
Article Keywords
Keyword Highlighting
Highlight selected keywords in the article text.
Timeline
| Received | August 25, 2025 |
| Accepted | September 24, 2025 |
| Published | December 28, 2025 |
licence
This license enables reusers to distribute, remix, adapt, and build upon the material in any medium or format for noncommercial purposes only, and only so long as attribution is given to the creator.